| Metric | Man Kendall Statistic | Normalized Test Statistic | p Value | Trend | Sens Slope |
|---|---|---|---|---|---|
| area | -0.2159 | -1.7509 | 0.0800 | no trend | -0.0044 |
| cai | -0.0909 | -0.7282 | 0.4665 | no trend | -0.0038 |
| circle | -0.0909 | -0.7282 | 0.4665 | no trend | -0.0038 |
| contig | -0.0909 | -0.7282 | 0.4665 | no trend | -0.0038 |
| core | -0.1667 | -1.3480 | 0.1777 | no trend | -0.0004 |
| enn | 0.4659 | 3.7961 | 0.0001 | increasing | 1.7304 |
| frac | -0.3826 | -3.1144 | 0.0018 | decreasing | -0.0004 |
| gyrate | -0.2652 | -2.1537 | 0.0313 | decreasing | -0.1367 |
| ncore | -0.1591 | -1.2860 | 0.1984 | no trend | -0.0013 |
| para | -0.1477 | -1.1931 | 0.2328 | no trend | 0.0000 |
| perim | -0.2614 | -2.1227 | 0.0338 | decreasing | -3.6115 |
| shape | -0.3106 | -2.5256 | 0.0116 | decreasing | -0.0037 |
| mean_mag | 0.0114 | 0.0775 | 0.9382 | no trend | 0.0355 |
| percent_area_dist | -0.3674 | -2.9904 | 0.0028 | decreasing | -0.0227 |
Historical Disturbance Trends: JOF Tuning (Recovery Threshold 0.75) Results
What’s Changed Since Last Time?
All results are now using an updated run of LT with the recovery threshold parameter set to 0.75. All other parameters are their default values
Man Kendall Statistics have been updated to actually be the MK Statistic
Parameters and Groups
LT Parameters
maxSegments: 6
spikeThreshold: 0.9
vertexCountOvershoot: 3
preventOneYearRecovery: true
recoveryThreshold: 0.75
pvalThreshold: 0.05
bestModelProportion: 0.75
minObservationsNeeded: 6
Size and Magnitude Thresholds
Minimum Magnitude: 50
Minimum Patch size: 0.27 ha (3 pixels)
Age Groups
Group 1: Aquistion prior to 1896
Group 2: Acquisition between 1897 and 1909
Group 3: Acquisition between 1910 and 1957
Group 4: Acquisition between 1958 and 1990
Man Kendall Test Results for Patch Level Metrics
This level of analysis includes all Forest Preserve land that has a completed UMP.
In changing to this set of parameters, we see a loss of a trend in mean magnitude and perimeter to area ratio, as well as new trends in radius of gyration and perimeter length. No metrics reversed the direction of trend.
Metric Plots
All patch metrics derived from the landscape metrics package in r. Descriptions of the patch metrics can be found here.
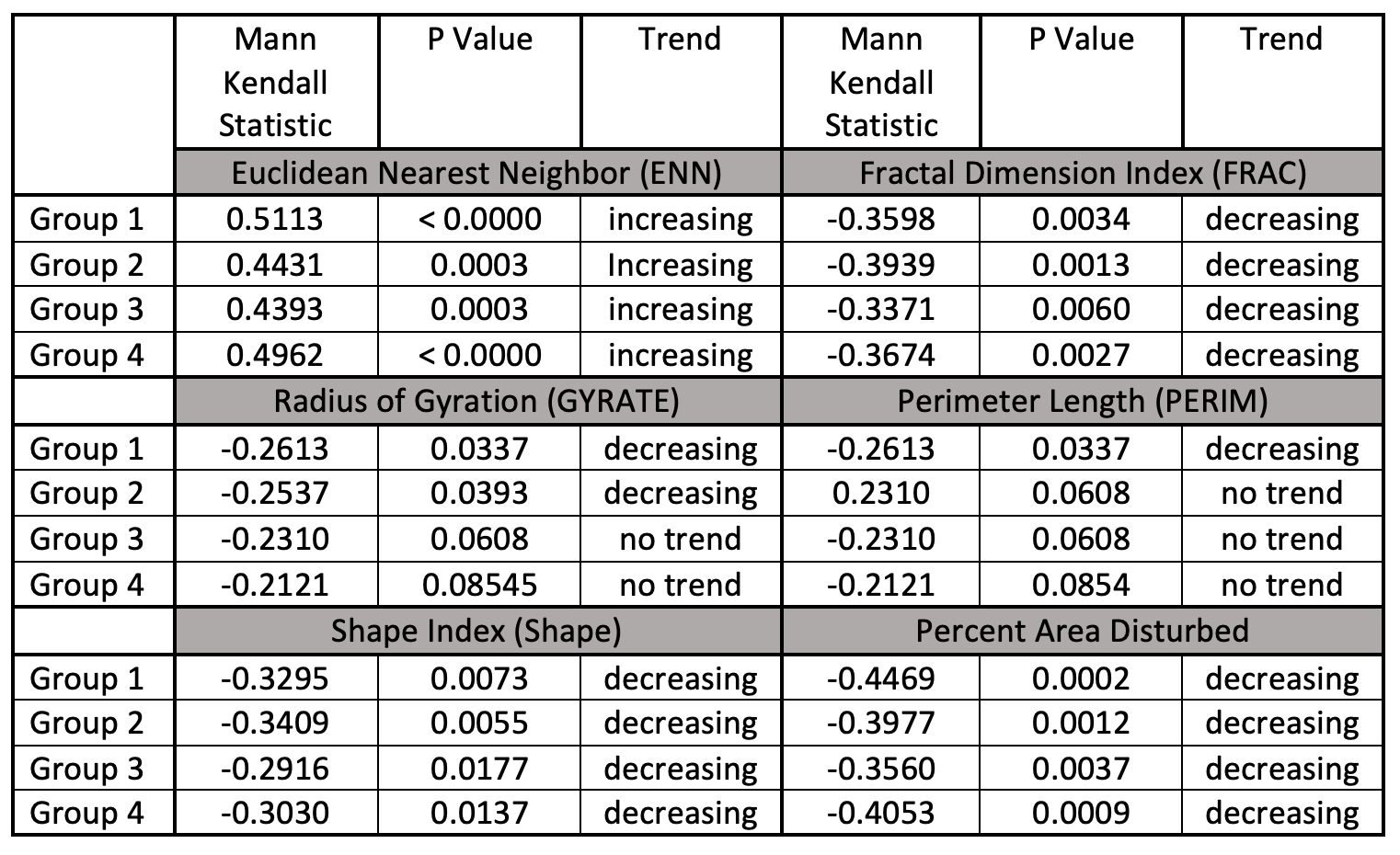
Age Group Mann Kendall Analysis
Metrics included in this table were selected because at least one group showed a significant trend for that metric. Excluded metrics showed no trends for any group.
Like in previous rounds of results, there were no metrics that showed trends in the group analysis that did not show trends in the overall analysis. Additionally, there again were no metrics that showed diverging trends in the group analysis. All groups that showed a trend matched the trend of the overall metric.
Metric Age Group ANOVAs
ANOVA for metric values by age group. Outliers are hidden for all plots (except core area metrics) for clarity.
Area
Analysis of Variance Table
Response: area
Df Sum Sq Mean Sq F value Pr(>F)
bin 3 206 68.721 1.9107 0.1254
Residuals 434971 15644737 35.967 Tukey multiple comparisons of means
95% family-wise confidence level
Fit: aov(formula = model)
$`grouped_patches$bin`
diff lwr upr p adj
2-1 0.002742793 -0.06403531 0.06952090 0.9995798
3-1 0.049089655 -0.01041688 0.10859619 0.1469003
4-1 0.036461065 -0.03035427 0.10327640 0.4980902
3-2 0.046346862 -0.02525643 0.11795015 0.3435136
4-2 0.033718272 -0.04406535 0.11150190 0.6811082
4-3 -0.012628589 -0.08426660 0.05900942 0.9690884CAI
CAI is the patch core area index, which is equal to the percentage of the patch that is not edge. For the vast majority of the patches analyzed here, all cells are edge cells (core = 0) so this metric is hugely zero inflated. This pattern is visible in all the core area metrics.
Analysis of Variance Table
Response: cai
Df Sum Sq Mean Sq F value Pr(>F)
bin 3 331 110.223 9.7229 2.065e-06 ***
Residuals 434971 4931024 11.336
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 Tukey multiple comparisons of means
95% family-wise confidence level
Fit: aov(formula = model)
$`grouped_patches$bin`
diff lwr upr p adj
2-1 0.01387420 -0.02361606 0.051364468 0.7773456
3-1 0.06823522 0.03482732 0.101643107 0.0000009
4-1 0.03497434 -0.00253682 0.072485508 0.0779499
3-2 0.05436101 0.01416181 0.094560210 0.0028825
4-2 0.02110014 -0.02256879 0.064769074 0.6004718
4-3 -0.03326087 -0.07347956 0.006957818 0.1452521Circle
Analysis of Variance Table
Response: circle
Df Sum Sq Mean Sq F value Pr(>F)
bin 3 0.1 0.047223 5.393 0.001042 **
Residuals 434971 3808.8 0.008756
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 Tukey multiple comparisons of means
95% family-wise confidence level
Fit: aov(formula = model)
$`grouped_patches$bin`
diff lwr upr p adj
2-1 -0.0003942335 -0.001436171 6.477037e-04 0.7654018
3-1 -0.0010065084 -0.001934987 -7.802935e-05 0.0274588
4-1 -0.0014763981 -0.002518916 -4.338800e-04 0.0015644
3-2 -0.0006122749 -0.001729499 5.049495e-04 0.4943229
4-2 -0.0010821646 -0.002295821 1.314914e-04 0.1001512
4-3 -0.0004698897 -0.001587656 6.478765e-04 0.7017970Contig
Analysis of Variance Table
Response: contig
Df Sum Sq Mean Sq F value Pr(>F)
bin 3 0.4 0.135510 13.094 1.515e-08 ***
Residuals 434971 4501.7 0.010349
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 Tukey multiple comparisons of means
95% family-wise confidence level
Fit: aov(formula = model)
$`grouped_patches$bin`
diff lwr upr p adj
2-1 -0.0002646962 -0.0013974561 0.0008680637 0.9319851
3-1 0.0019669311 0.0009575192 0.0029763430 0.0000033
4-1 -0.0005377883 -0.0016711797 0.0005956030 0.6147318
3-2 0.0022316273 0.0010170176 0.0034462370 0.0000140
4-2 -0.0002730922 -0.0015925391 0.0010463548 0.9513767
4-3 -0.0025047194 -0.0037199181 -0.0012895208 0.0000007Core
Like CAI above, this metric measures the total area that is core area in a patch (in ha). For the vast majority of the patches analyzed here, all cells are edge cells (core = 0) so this metric is hugely zero inflated.
0% 25% 50% 75% 100%
0.00 0.00 0.00 0.00 567.54 Analysis of Variance Table
Response: core
Df Sum Sq Mean Sq F value Pr(>F)
bin 3 36 12.0896 2.0152 0.1094
Residuals 434971 2609459 5.9992 Tukey multiple comparisons of means
95% family-wise confidence level
Fit: aov(formula = model)
$`grouped_patches$bin`
diff lwr upr p adj
2-1 0.005688892 -0.021583622 0.03296141 0.9503073
3-1 0.019380183 -0.004922585 0.04368295 0.1703037
4-1 0.020347221 -0.006940497 0.04763494 0.2213767
3-2 0.013691291 -0.015551852 0.04293443 0.6250540
4-2 0.014658329 -0.017108894 0.04642555 0.6360814
4-3 0.000967038 -0.028290285 0.03022436 0.9997807ENN
Analysis of Variance Table
Response: enn
Df Sum Sq Mean Sq F value Pr(>F)
bin 3 124924 41641 17.811 1.486e-11 ***
Residuals 434971 1016969021 2338
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 Tukey multiple comparisons of means
95% family-wise confidence level
Fit: aov(formula = model)
$`grouped_patches$bin`
diff lwr upr p adj
2-1 0.3569120 -0.18148649 0.8953104 0.3220075
3-1 0.5741742 0.09440282 1.0539457 0.0113340
4-1 1.5192864 0.98058781 2.0579850 0.0000000
3-2 0.2172623 -0.36003925 0.7945638 0.7683395
4-2 1.1623744 0.53524398 1.7895049 0.0000114
4-3 0.9451122 0.36753074 1.5226936 0.0001541Frac
Analysis of Variance Table
Response: enn
Df Sum Sq Mean Sq F value Pr(>F)
bin 3 124924 41641 17.811 1.486e-11 ***
Residuals 434971 1016969021 2338
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 Tukey multiple comparisons of means
95% family-wise confidence level
Fit: aov(formula = model)
$`grouped_patches$bin`
diff lwr upr p adj
2-1 0.3569120 -0.18148649 0.8953104 0.3220075
3-1 0.5741742 0.09440282 1.0539457 0.0113340
4-1 1.5192864 0.98058781 2.0579850 0.0000000
3-2 0.2172623 -0.36003925 0.7945638 0.7683395
4-2 1.1623744 0.53524398 1.7895049 0.0000114
4-3 0.9451122 0.36753074 1.5226936 0.0001541Gyrate
Analysis of Variance Table
Response: gyrate
Df Sum Sq Mean Sq F value Pr(>F)
bin 3 11641 3880.5 5.1865 0.001396 **
Residuals 434971 325436987 748.2
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 Tukey multiple comparisons of means
95% family-wise confidence level
Fit: aov(formula = model)
$`grouped_patches$bin`
diff lwr upr p adj
2-1 -0.16813537 -0.47270276 0.13643203 0.4878567
3-1 0.21496498 -0.05643759 0.48636756 0.1751939
4-1 -0.23511373 -0.53985091 0.06962345 0.1947045
3-2 0.38310035 0.05652584 0.70967486 0.0137533
4-2 -0.06697837 -0.42174068 0.28778395 0.9624461
4-3 -0.45007872 -0.77681158 -0.12334586 0.0022722Ncore
The third core area metric, ncore represents the number of core areas in the patch. As in the other core area metrics this one is again heavily zero inflated.
Analysis of Variance Table
Response: ncore
Df Sum Sq Mean Sq F value Pr(>F)
bin 3 22 7.4172 3.0282 0.02819 *
Residuals 434971 1065390 2.4493
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 Tukey multiple comparisons of means
95% family-wise confidence level
Fit: aov(formula = model)
$`grouped_patches$bin`
diff lwr upr p adj
2-1 -3.401856e-05 -0.0174602807 0.01739224 1.0000000
3-1 1.589351e-02 0.0003648181 0.03142220 0.0425102
4-1 1.155515e-02 -0.0058808229 0.02899113 0.3222712
3-2 1.592753e-02 -0.0027579051 0.03461296 0.1259968
4-2 1.158917e-02 -0.0087090646 0.03188741 0.4577259
4-3 -4.338354e-03 -0.0230328450 0.01435614 0.9332594Para
Analysis of Variance Table
Response: para
Df Sum Sq Mean Sq F value Pr(>F)
bin 3 0.012 0.0038463 11.745 1.088e-07 ***
Residuals 434971 142.448 0.0003275
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 Tukey multiple comparisons of means
95% family-wise confidence level
Fit: aov(formula = model)
$`grouped_patches$bin`
diff lwr upr p adj
2-1 4.727698e-05 -0.0001542248 0.0002487787 0.9312262
3-1 -3.436630e-04 -0.0005232229 -0.0001641031 0.0000052
4-1 5.378769e-05 -0.0001478264 0.0002554018 0.9027153
3-2 -3.909400e-04 -0.0006070016 -0.0001748783 0.0000199
4-2 6.510716e-06 -0.0002282000 0.0002412214 0.9998703
4-3 3.974507e-04 0.0001812843 0.0006136171 0.0000138Perim
Analysis of Variance Table
Response: perim
Df Sum Sq Mean Sq F value Pr(>F)
bin 3 2.2021e+07 7340268 1.6879 0.1672
Residuals 434971 1.8916e+12 4348717 Tukey multiple comparisons of means
95% family-wise confidence level
Fit: aov(formula = model)
$`grouped_patches$bin`
diff lwr upr p adj
2-1 -3.422746 -26.642663 19.79717 0.9815101
3-1 15.490641 -5.200823 36.18210 0.2181717
4-1 5.742728 -17.490134 28.97559 0.9207400
3-2 18.913387 -5.984331 43.81111 0.2066687
4-2 9.165474 -17.881254 36.21220 0.8200848
4-3 -9.747913 -34.657704 15.16188 0.7462928Shape
Analysis of Variance Table
Response: shape
Df Sum Sq Mean Sq F value Pr(>F)
bin 3 5 1.8201 5.4576 0.000951 ***
Residuals 434971 145064 0.3335
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 Tukey multiple comparisons of means
95% family-wise confidence level
Fit: aov(formula = model)
$`grouped_patches$bin`
diff lwr upr p adj
2-1 -0.0043150511 -0.010745335 0.002115233 0.3111140
3-1 -0.0006042748 -0.006334355 0.005125806 0.9930511
4-1 -0.0094357660 -0.015869635 -0.003001897 0.0009472
3-2 0.0037107763 -0.003184141 0.010605693 0.5102576
4-2 -0.0051207150 -0.012610756 0.002369327 0.2946350
4-3 -0.0088314912 -0.015729751 -0.001933231 0.0055509Mean Magnitude
Analysis of Variance Table
Response: mean_mag
Df Sum Sq Mean Sq F value Pr(>F)
bin 3 293115 97705 40.307 < 2.2e-16 ***
Residuals 434971 1054366469 2424
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 Tukey multiple comparisons of means
95% family-wise confidence level
Fit: aov(formula = model)
$`grouped_patches$bin`
diff lwr upr p adj
2-1 -0.1068570 -0.6550655 0.4413514 0.9589077
3-1 0.1259940 -0.3625192 0.6145072 0.9111208
4-1 -2.1097487 -2.6582628 -1.5612346 0.0000000
3-2 0.2328511 -0.3549693 0.8206715 0.7390950
4-2 -2.0028917 -2.6414489 -1.3643344 0.0000000
4-3 -2.2357428 -2.8238481 -1.6476374 0.0000000Summary
With the exception of the area and core metrics, at least one age group is significantly different for all metrics. There are no metrics where each age group was significantly different from all other groups. There are no clear patterns within metric types (shape, area/edge, core area).
Outlier Year ANOVAs
ANOVA by age groups for years with outlier values in specific metrics. Outliers again removed for clarity.
Mean Disturbance Magnitude Outliers
1995
Analysis of Variance Table
Response: mean_mag
Df Sum Sq Mean Sq F value Pr(>F)
bin 3 101706 33902 10.873 3.941e-07 ***
Residuals 15122 47151532 3118
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 Tukey multiple comparisons of means
95% family-wise confidence level
Fit: aov(formula = model)
$`year_patches$bin`
diff lwr upr p adj
2-1 2.915925 -0.5399446 6.371794 0.1322957
3-1 1.396193 -1.5901035 4.382490 0.6260488
4-1 6.981866 3.7508511 10.212880 0.0000002
3-2 -1.519731 -5.1011828 2.061720 0.6955256
4-2 4.065941 0.2780285 7.853853 0.0297134
4-3 5.585672 2.2206733 8.950671 0.00011832007
Analysis of Variance Table
Response: mean_mag
Df Sum Sq Mean Sq F value Pr(>F)
bin 3 74567 24855.6 4.5969 0.003298 **
Residuals 1420 7678055 5407.1
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 Tukey multiple comparisons of means
95% family-wise confidence level
Fit: aov(formula = model)
$`year_patches$bin`
diff lwr upr p adj
2-1 -10.340278 -25.43052 4.749968 0.2920190
3-1 -16.686987 -28.93591 -4.438065 0.0026599
4-1 -2.796400 -18.40756 12.814766 0.9675262
3-2 -6.346709 -21.02748 8.334063 0.6822233
4-2 7.543878 -10.04013 25.127890 0.6874547
4-3 13.890587 -1.32513 29.106305 0.08790172021
Analysis of Variance Table
Response: mean_mag
Df Sum Sq Mean Sq F value Pr(>F)
bin 3 14468 4822.6 2.6842 0.045 *
Residuals 9641 17321665 1796.7
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 Tukey multiple comparisons of means
95% family-wise confidence level
Fit: aov(formula = model)
$`year_patches$bin`
diff lwr upr p adj
2-1 0.5176050 -2.656541 3.6917513 0.9752491
3-1 -0.2902162 -3.219491 2.6390590 0.9942160
4-1 -2.8420742 -5.870666 0.1865176 0.0750128
3-2 -0.8078212 -4.183014 2.5673714 0.9273474
4-2 -3.3596793 -6.821419 0.1020603 0.0609324
4-3 -2.5518581 -5.790544 0.6868282 0.1789431